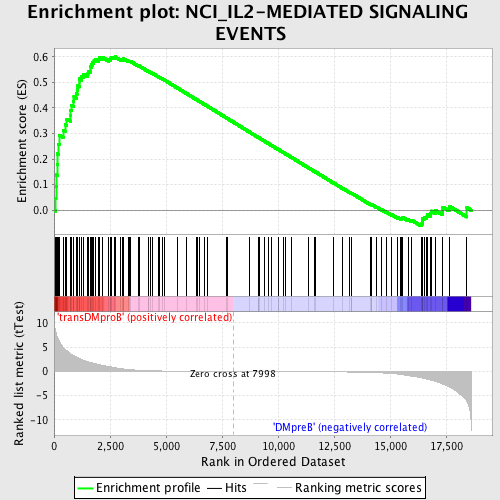
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | NCI_IL2-MEDIATED SIGNALING EVENTS |
| Enrichment Score (ES) | 0.59962434 |
| Normalized Enrichment Score (NES) | 1.5434394 |
| Nominal p-value | 0.006756757 |
| FDR q-value | 0.24059758 |
| FWER p-Value | 0.969 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SOCS2 | 5694 | 65 | 8.301 | 0.0463 | Yes | ||
| 2 | MAPKAPK2 | 13838 | 85 | 7.980 | 0.0931 | Yes | ||
| 3 | JAK3 | 9198 4936 | 107 | 7.705 | 0.1382 | Yes | ||
| 4 | TNFAIP3 | 19810 | 138 | 7.145 | 0.1794 | Yes | ||
| 5 | CISH | 8743 | 157 | 6.972 | 0.2203 | Yes | ||
| 6 | STAT1 | 3936 5524 | 190 | 6.574 | 0.2580 | Yes | ||
| 7 | ELF1 | 21948 | 231 | 6.237 | 0.2932 | Yes | ||
| 8 | MAPK14 | 23313 | 429 | 4.797 | 0.3114 | Yes | ||
| 9 | NFKB2 | 23810 | 491 | 4.513 | 0.3351 | Yes | ||
| 10 | STAT3 | 5525 9906 | 569 | 4.247 | 0.3564 | Yes | ||
| 11 | MAPK1 | 1642 11167 | 721 | 3.643 | 0.3701 | Yes | ||
| 12 | STAT5A | 20664 | 727 | 3.634 | 0.3917 | Yes | ||
| 13 | LCK | 15746 | 786 | 3.491 | 0.4095 | Yes | ||
| 14 | RASA1 | 10174 | 860 | 3.276 | 0.4252 | Yes | ||
| 15 | MAP3K14 | 11998 | 884 | 3.224 | 0.4433 | Yes | ||
| 16 | JAK1 | 15827 | 996 | 2.955 | 0.4550 | Yes | ||
| 17 | CALM3 | 8682 | 1025 | 2.875 | 0.4707 | Yes | ||
| 18 | CCND3 | 4489 4490 | 1057 | 2.813 | 0.4859 | Yes | ||
| 19 | DOK2 | 354 | 1136 | 2.652 | 0.4976 | Yes | ||
| 20 | TRAF6 | 5797 14940 | 1138 | 2.646 | 0.5134 | Yes | ||
| 21 | MAPK9 | 1233 20903 1383 | 1236 | 2.449 | 0.5229 | Yes | ||
| 22 | KRAS | 9247 | 1320 | 2.284 | 0.5321 | Yes | ||
| 23 | PTK2B | 21776 | 1509 | 1.984 | 0.5339 | Yes | ||
| 24 | GRB2 | 20149 | 1534 | 1.943 | 0.5442 | Yes | ||
| 25 | NFKB1 | 15160 | 1611 | 1.847 | 0.5512 | Yes | ||
| 26 | RELA | 23783 | 1612 | 1.845 | 0.5622 | Yes | ||
| 27 | JUN | 15832 | 1670 | 1.759 | 0.5697 | Yes | ||
| 28 | ARRB2 | 20806 | 1715 | 1.707 | 0.5776 | Yes | ||
| 29 | SOCS3 | 20131 | 1779 | 1.638 | 0.5840 | Yes | ||
| 30 | CALM2 | 8681 | 1832 | 1.596 | 0.5908 | Yes | ||
| 31 | STAM | 2912 15117 | 1995 | 1.404 | 0.5904 | Yes | ||
| 32 | CDK2 | 3438 3373 19592 3322 | 2032 | 1.374 | 0.5967 | Yes | ||
| 33 | BIRC2 | 4397 4398 | 2142 | 1.276 | 0.5985 | Yes | ||
| 34 | RAN | 5356 9691 | 2444 | 1.000 | 0.5882 | Yes | ||
| 35 | RAC1 | 16302 | 2500 | 0.969 | 0.5911 | Yes | ||
| 36 | MAPK3 | 6458 11170 | 2529 | 0.941 | 0.5952 | Yes | ||
| 37 | CDK6 | 16600 | 2567 | 0.903 | 0.5986 | Yes | ||
| 38 | MYB | 3344 3355 19806 | 2693 | 0.791 | 0.5966 | Yes | ||
| 39 | FOS | 21202 | 2723 | 0.766 | 0.5996 | Yes | ||
| 40 | RAF1 | 17035 | 2978 | 0.564 | 0.5893 | No | ||
| 41 | MYC | 22465 9435 | 3045 | 0.531 | 0.5889 | No | ||
| 42 | CCND2 | 16987 | 3097 | 0.503 | 0.5892 | No | ||
| 43 | MAP2K1 | 19082 | 3106 | 0.498 | 0.5917 | No | ||
| 44 | SP1 | 9852 | 3303 | 0.399 | 0.5835 | No | ||
| 45 | TERT | 21604 | 3369 | 0.372 | 0.5822 | No | ||
| 46 | NOD2 | 6384 | 3419 | 0.353 | 0.5817 | No | ||
| 47 | SMPD1 | 18145 2658 | 3760 | 0.240 | 0.5648 | No | ||
| 48 | CYLD | 18532 | 3798 | 0.230 | 0.5642 | No | ||
| 49 | RPS6KB1 | 7815 1207 13040 | 4207 | 0.147 | 0.5430 | No | ||
| 50 | PRF1 | 20011 | 4217 | 0.145 | 0.5434 | No | ||
| 51 | BCL2L1 | 4440 2930 8652 | 4316 | 0.132 | 0.5389 | No | ||
| 52 | TNF | 23004 | 4390 | 0.120 | 0.5356 | No | ||
| 53 | BCL10 | 15397 | 4641 | 0.092 | 0.5227 | No | ||
| 54 | SOS1 | 5476 | 4708 | 0.086 | 0.5196 | No | ||
| 55 | GAB2 | 1821 18184 2025 | 4834 | 0.077 | 0.5133 | No | ||
| 56 | PIK3R1 | 3170 | 4927 | 0.072 | 0.5088 | No | ||
| 57 | FBXW11 | 20926 | 5500 | 0.046 | 0.4782 | No | ||
| 58 | LTA | 23003 | 5889 | 0.035 | 0.4574 | No | ||
| 59 | ERC1 | 1013 17021 995 1136 | 6369 | 0.025 | 0.4317 | No | ||
| 60 | INSL3 | 9181 | 6415 | 0.024 | 0.4294 | No | ||
| 61 | RIPK2 | 2528 15935 | 6494 | 0.023 | 0.4253 | No | ||
| 62 | ATM | 2976 19115 | 6719 | 0.018 | 0.4133 | No | ||
| 63 | FOXO3 | 19782 3402 | 6840 | 0.016 | 0.4069 | No | ||
| 64 | FASLG | 13789 | 7673 | 0.004 | 0.3620 | No | ||
| 65 | PIK3CA | 9562 | 7760 | 0.003 | 0.3574 | No | ||
| 66 | STAT5B | 20222 | 8729 | -0.010 | 0.3051 | No | ||
| 67 | BCL2 | 8651 3928 13864 4435 981 4062 13863 4027 | 9115 | -0.015 | 0.2844 | No | ||
| 68 | SHC1 | 9813 9812 5430 | 9137 | -0.016 | 0.2834 | No | ||
| 69 | FRAP1 | 2468 15991 | 9178 | -0.016 | 0.2813 | No | ||
| 70 | IL4 | 9174 | 9385 | -0.019 | 0.2703 | No | ||
| 71 | FYN | 3375 3395 20052 | 9588 | -0.022 | 0.2595 | No | ||
| 72 | PTPN11 | 5326 16391 9660 | 9715 | -0.024 | 0.2528 | No | ||
| 73 | PRKCE | 9575 | 10004 | -0.029 | 0.2374 | No | ||
| 74 | IRS1 | 4925 | 10031 | -0.029 | 0.2362 | No | ||
| 75 | IL2 | 15354 | 10229 | -0.032 | 0.2258 | No | ||
| 76 | IKBKG | 2570 2562 4908 | 10350 | -0.035 | 0.2195 | No | ||
| 77 | CCNA2 | 15357 | 10583 | -0.038 | 0.2072 | No | ||
| 78 | SRC | 5507 | 11343 | -0.054 | 0.1665 | No | ||
| 79 | CALM1 | 21184 | 11604 | -0.060 | 0.1528 | No | ||
| 80 | PRKCA | 20174 | 11681 | -0.062 | 0.1490 | No | ||
| 81 | IFNG | 19869 | 12487 | -0.093 | 0.1061 | No | ||
| 82 | XPO1 | 4172 | 12880 | -0.112 | 0.0856 | No | ||
| 83 | AKT1 | 8568 | 13190 | -0.131 | 0.0697 | No | ||
| 84 | STAM2 | 7095 2845 | 13282 | -0.138 | 0.0656 | No | ||
| 85 | RHOA | 8624 4409 4410 | 13287 | -0.138 | 0.0662 | No | ||
| 86 | MAPK11 | 2264 9618 22163 | 14126 | -0.222 | 0.0222 | No | ||
| 87 | TNFRSF1A | 1181 10206 | 14170 | -0.228 | 0.0213 | No | ||
| 88 | FOXP3 | 9804 5426 | 14182 | -0.231 | 0.0221 | No | ||
| 89 | PPP2R5D | 962 1523 22957 10139 | 14395 | -0.263 | 0.0122 | No | ||
| 90 | REL | 9716 | 14615 | -0.310 | 0.0022 | No | ||
| 91 | RELB | 17942 | 14859 | -0.372 | -0.0087 | No | ||
| 92 | IL2RG | 24096 | 15069 | -0.442 | -0.0173 | No | ||
| 93 | PRKCZ | 5260 | 15328 | -0.546 | -0.0280 | No | ||
| 94 | SYK | 21636 | 15479 | -0.643 | -0.0322 | No | ||
| 95 | UBE2D3 | 7253 | 15527 | -0.667 | -0.0308 | No | ||
| 96 | UGCG | 16197 | 15558 | -0.683 | -0.0283 | No | ||
| 97 | MAP2K2 | 19933 | 15807 | -0.865 | -0.0365 | No | ||
| 98 | IL2RB | 22219 | 15966 | -0.989 | -0.0391 | No | ||
| 99 | NFKBIA | 21065 | 16384 | -1.316 | -0.0538 | No | ||
| 100 | RPS6 | 9757 | 16428 | -1.358 | -0.0479 | No | ||
| 101 | NRAS | 5191 | 16449 | -1.395 | -0.0406 | No | ||
| 102 | MAPK8 | 6459 | 16452 | -1.403 | -0.0323 | No | ||
| 103 | IKBKB | 4907 | 16520 | -1.483 | -0.0271 | No | ||
| 104 | HSP90AA1 | 4883 4882 9131 | 16644 | -1.583 | -0.0242 | No | ||
| 105 | MALT1 | 6274 | 16663 | -1.599 | -0.0156 | No | ||
| 106 | HRAS | 4868 | 16802 | -1.753 | -0.0125 | No | ||
| 107 | BCL3 | 8654 | 16826 | -1.782 | -0.0031 | No | ||
| 108 | SOCS1 | 4522 | 17035 | -2.077 | -0.0019 | No | ||
| 109 | E2F1 | 14384 | 17346 | -2.611 | -0.0030 | No | ||
| 110 | IL2RA | 4918 | 17354 | -2.627 | 0.0124 | No | ||
| 111 | CHUK | 23665 | 17633 | -3.139 | 0.0162 | No | ||
| 112 | SGMS1 | 23698 | 18426 | -6.143 | 0.0103 | No |

